Seminar “DNA and protein translocation through glass nanopores”
Date
Thursday, 7 June 2016
Time
3:00 pm
Place
University of Barcelona
Faculty of Physics Building
Room V12M, 1st floor
Speaker
Dr Ulrich F. Keyser, Cavendish Laboratory, University of Cambridge (United Kingdom)
Abstract
Solid-state nanopores are single molecule sensors for detection and analysis of biomolecules like DNA, RNA and proteins. The idea is as simple as intriguing since the sensing relies on measuring the changes in ionic current as a macromolecule passes.
First, we will discuss our recent results on DNA translocations. We observe an entropic barrier limited, length dependent translocation rate at 4M LiCl salt concentration and a drift-dominated, length independent translocation rate at 1M KCl salt concentration. These observations are described by a unifying convection-diffusion equation which includes the contribution of an entropic barrier for polymer entry [1].
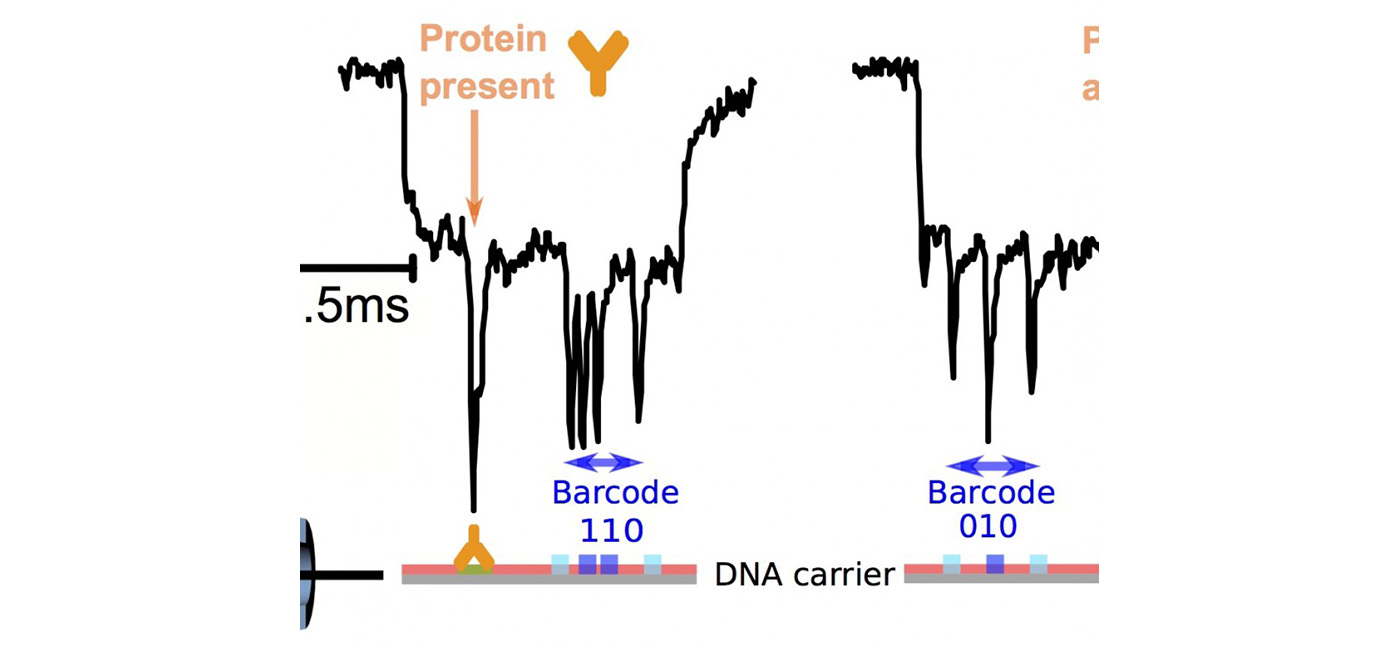
Based on our understanding of DNA translocation, we introduce DNA carriers that have specific protein binding sites to control protein translocation. Protein detection down to the single protein level is achieved, allowing for identification a single protein species within complex mixture [2]. Finally, the method is extended to multiplexed protein sensing [3].
References:
[1] N. A. W. Bell, M. Muthukumar, and U. F. Keyser. Translocation frequency of double-stranded DNA through a solid-state nanopore. Phys. Rev. E, 93:022401, 2016
[2] N. A. W. Bell and U. F. Keyser. Specific Protein Detection using Designed DNA Carriers and Nanopores. JACS, 137(5):2035-2041, 2015
[3] N. A. W. Bell and U. F. Keyser. Digitally encoded DNA nanostructures for multiplexed, singlemolecule protein sensing with nanopores. Nature Nanotechnology, published online, 2016


