Nucleic acids biophysics
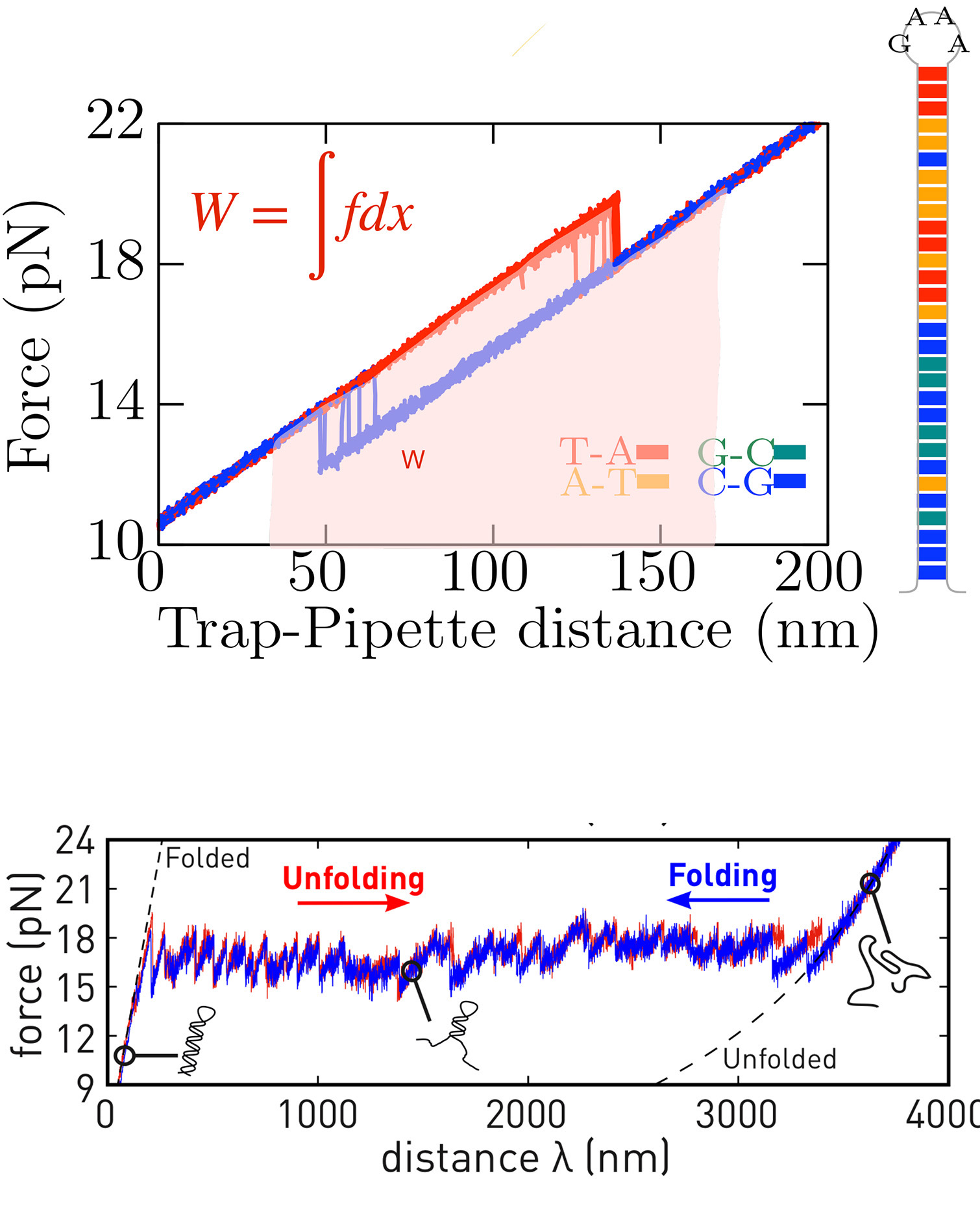
Much of our laboratory’s work focuses on accurately determining the thermodynamic properties of nucleic acids—both DNA and RNA—using force spectroscopy and optical tweezers. We have pioneered the combination of single-molecule DNA unzipping experiments with advanced tools from statistical physics to extract the nearest-neighbor free energy parameters for base pairing. This approach has achieved the remarkable accuracy of 0.1 kcal/mol in measurements at room temperature (25 °C) [1,2].
By extending these methods through the framework of stochastic thermodynamics, we have also characterized RNA hybridization thermodynamics [3]. Compared to DNA, RNA exhibits pronounced irreversibility effects, which we attribute to a more rugged energy landscape and additional interactions involving the 2′-OH group in ribose with nucleotides, ions, and water (see our Cold RNA research line).
To further this work, we developed a temperature-controlled optical tweezers system [4], enabling us to measure enthalpy, entropy, and the lesser-known heat capacity changes of DNA at single base-pair resolution [5]. This innovation lays the foundation for the emerging field of calorimetric force spectroscopy. Notably, a refined adaptation of the Clausius-Clapeyron equation to force measurements has allowed us to determine the heat capacity changes of individual nearest-neighbor base pairs with unprecedented accuracy.
[1] J. M. Huguet, C. V. Bizarro, N. Forns, S. B. Smith, C. Bustamante and F. Ritort, Single-molecule derivation of salt dependent base-pair free energies in DNA, Proceedings of the National Academy of Sciences, 107 (2010) 15431-15436
[2] J. M. Huguet, M. Ribezzi-Crivellari, C. V. Bizarro and F. Ritort Derivation of nearest-neighbor DNA parameters in magnesium from single-molecule experiments, Nucleic Acids Research 45 (2017) 12921-12931
[3] P. Rissone, C. V. Bizarro, F. Ritort, Stem loop formation drives RNA folding in mechanical unzipping experiments, Proceedings of the National Academy of Sciences 119 (3) e2025575119 (2022)
[4] S. de Lorenzo, M. Ribezzi-Crivellari, R. Arias-Gonzalez, S.B. Smith and F. Ritort, A Temperature-Jump Optical Trap for Single-Molecule Manipulation, Biophysical Journal, 108 (2015) 2854-2864
[5] P. Rissone, M. Rico-Pasto, S. B. Smith and F. Ritort, DNA Calorimetric Force Spectroscopy at Single Base Pair Resolution, Nature Communications 16 (2025) 2688
[6] J. Camunas-Soler, M. Manosas, S. Frutos, J. Tulla-Puche, F. Albericio and F. Ritort, Single-molecule kinetics and footprinting of DNA bis-intercalation: the paradigmatic case of Thiocoraline, Nucleic Acids Research, 43 (2015) 2767-2779
[7] M. Manosas, J. Camunas-Soler, V. Croquette and F. Ritort, Single molecule high-throughput footprinting of small and large DNA ligands, Nature Communications 8 (2017) 304.