Molecular folding
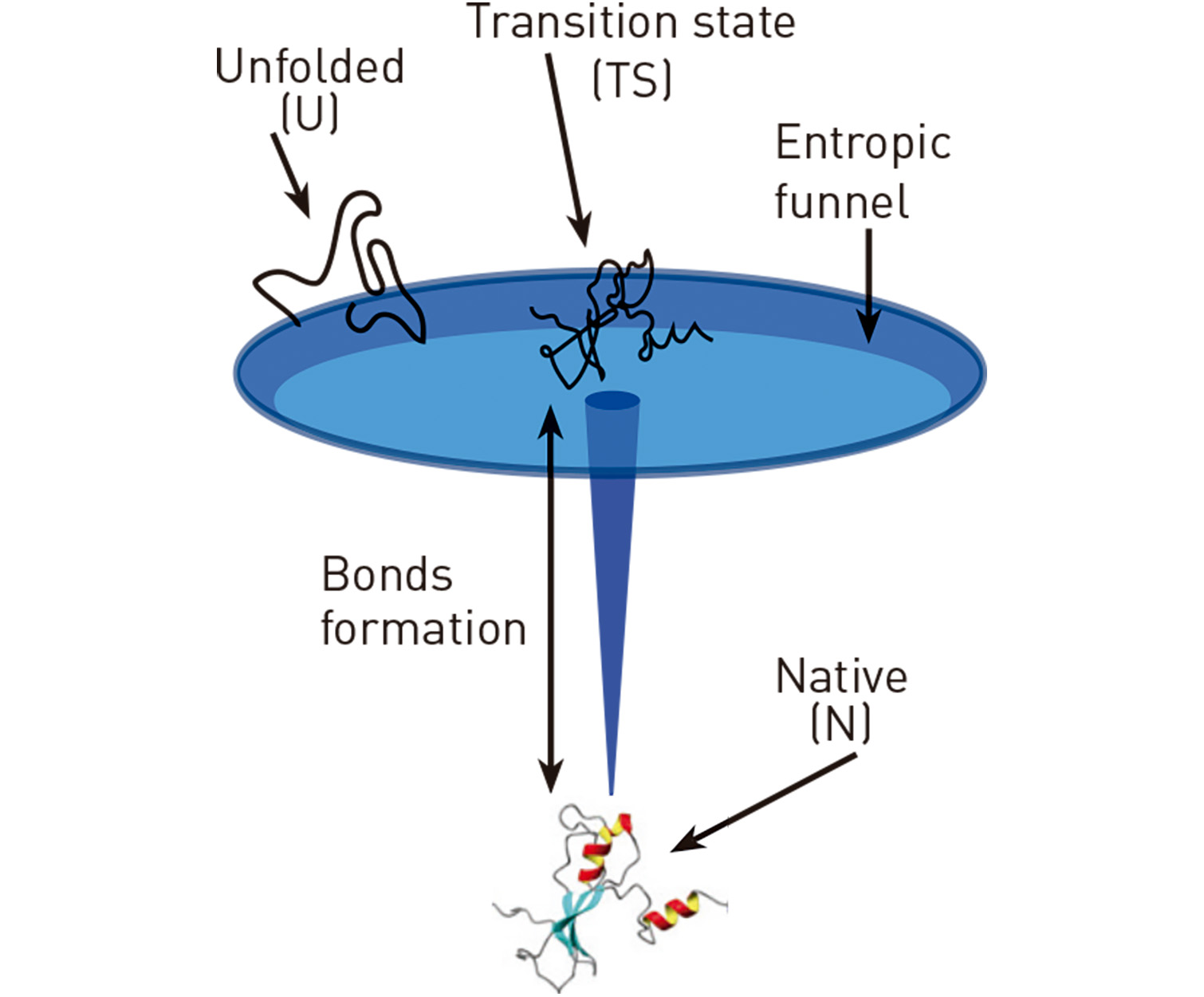
A fascinating non-equilibrium process in nature is molecular folding, or how molecules fold into their native functional three-dimensional structure. According to the Levinthal paradox, folding into the native structure amounts to finding a needle in a haystack. For a random search, successful folding would last the universe’s age. How do molecules circumvent Levinthal’s paradox and fold in finite times? Our studies of protein barnase [1,2] have revealed that folding occurs in two steps: first, the unfolded chain collapses into a low-configurational entropy transition state that is structurally similar to the native without stabilized bonds; next, the chain collapses into the lowest energy native structure. The folding landscape has the shape of a funnel, a result that we also demonstrated in DNA hairpins [3]. Molecular folding is strongly affected by cations, such as magnesium ions, that strongly influence RNA folding [4,5].
An exciting endeavor over the past years has been the discovery that helix-coil models can be applied to study conformational transitions in single-stranded DNA and RNA, such as the formation of non-specific secondary structure [6] and the stacking-unstacking transition [7], measuring the stacking energies with an unmatched 0.1 kcal/mol accuracy.
With force techniques, it is possible to manipulate single molecules while monitoring how they fold and unfold by repeatedly. We introduced the continuous effective barrier approach to reconstruct the folding energy landscape from kinetics measurements, which is especially useful for DNA and RNA hairpins, where the number of unzipped base pairs defines a one-dimensional (1D) reaction coordinate [8,9]. DNA unzipping, the process by which the two strands of the double helix are pulled apart, exhibits many intermediates [10]. Unzipping is a fracture process that has permitted the experimental test of the renormalization group flow equations of random field theories, and the measurement of critical exponents at the single molecule level for the first time [11].
[1] A. Alemany, B. Rey-Serra, S. Frutos, C. Cecconi, and F. Ritort, Mechanical Folding and Unfolding of Protein Barnase at the Single-Molecule Level, Biophysical Journal, 110 (2016) 63-74
[2] M. Rico-Pasto, A. Zaltron, S. Davies, S. Frutos and F. Ritort, Molten-globule like transition state of protein barnase measured with calorimetric force spectroscopy, Proceedings of the National Academy of Sciences 119 (11) e2112382119 (2022)
[3] M. Rico-Pasto, M. Ribezzi-Crivellari, F. Ritort, Temperature-dependent funnel-like DNA folding landscapes, under review in Nucleic Acids Research
[4] C. V. Bizarro, A. Alemany and F. Ritort, Non-specific binding of Na+ and Mg2+ to RNA determined by force spectroscopy methods, Nucleic Acids Research, 40 (2012) 6922-6935
[5] A. M. Monge., I. Pastor, C. Bustamante, M. Manosas and F. Ritort, Measurement of the Specific and Non-specific Binding Energies of Mg2+ to RNA, Biophysical Journal 21 (2022) 3010-3022
[6] X. Viader-Godoy, C. R. Pulido, B. Ibarra, M. Manosas and F. Ritort, Cooperativity-dependent folding of single stranded DNA, Physical Review X 11 (2021) 031037
[7] X. Viader-Godoy, M. Manosas and F. Ritort, Stacking correlation length in single-stranded DNA, Nucleic Acids Research 52 (2017) 13243-13254
[8] A. Alemany and F. Ritort, Force-Dependent Folding and Unfolding Kinetics in DNA Hairpins Reveals Transition State Displacements Along a Single Pathway, The Journal of Physical Chemistry Letters 8 (2017) 895–900
[9] M. Rico-Pasto, A. Alemany, and F. Ritort, Force-Dependent Folding Kinetics of Single Molecules with Multiple Intermediates and Pathways, J. Phys. Chem. Lett. 13, 4 (2022) 1025–1032
[10] J. M. Huguet, N. Forns and F. Ritort, Statistical properties of metastable intermediates in DNA unzipping, Physical Review Letters, 103 (2009) 248106
[11] C. ter Burg, P. Rissone, M. Rico-Pasto, F. Ritort, K. J. Wiese, Experimental test of Sinai’s model in DNA unzipping, Physical Review Letters 130 (2023) 208401