Seminar “RNA structural biology & bioinformatics”
Date
Thursday, 24 November 2016
Time
11:30 am
Place
University of Barcelona
Faculty of Physics Building
Room 3.20, 3rd floor
Speaker
Dr Eric Westhof, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire du CNRS (France)
Abstract
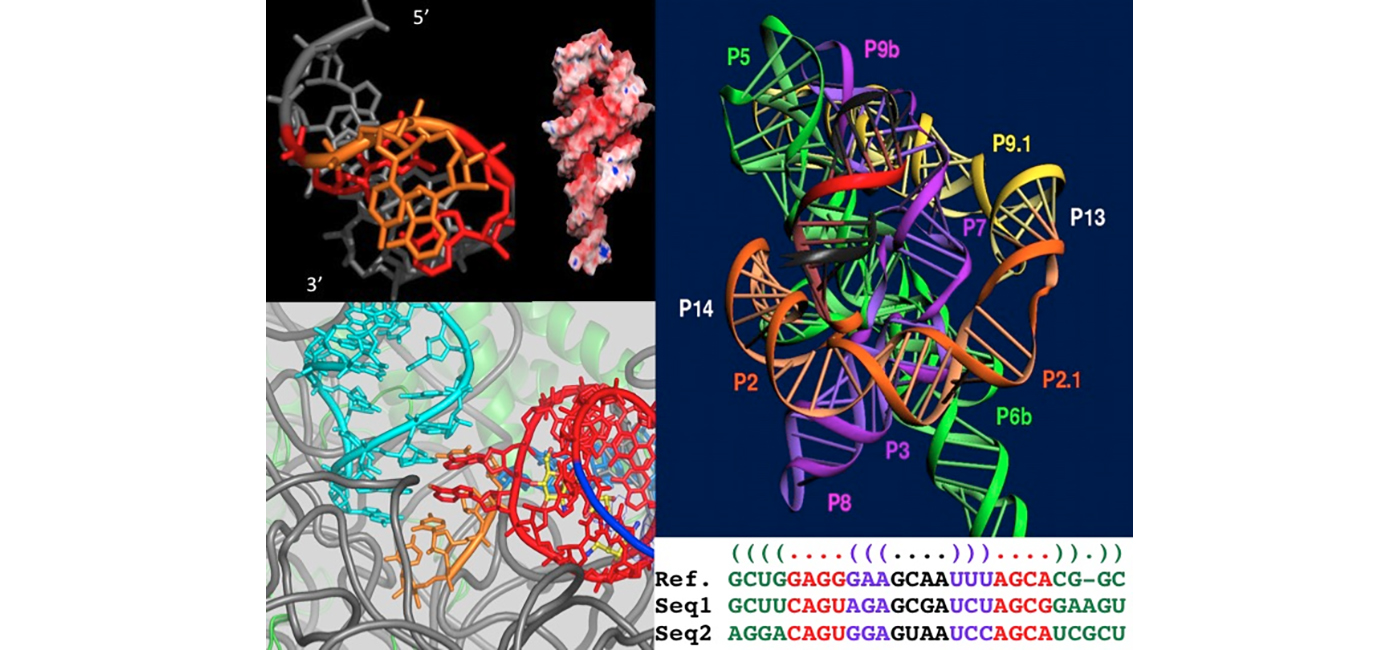
RNA modules are intrinsic to RNA architecture are therefore disconnected from a biological function specifically attached to a RNA sequence. RNA modules occur in all kingdoms of life and in structured RNAs with diverse functions. Because of chemical and geometrical constraints, isostericity between non-Watson-Crick pairs is restricted and this leads to higher sequence conservation in RNA modules with, consequently, greater difficulties in extracting 3D information from sequence analysis. The program RMDetect allows the identification of RNA modules in RNA sequences.
The understanding of the recognition principles of RNA binding to proteins is necessary to predict the binding interfaces. A simple score to predict RNA binding probabilities based on a combination of protein sequences and structures will be described. The prediction score is based on physico-chemical and evolutionary principles. RNA binding residues are accessible on protein surface, tend to be positively charged and are highly conserved in sequence. The derived RBscore is a combination of residue accessibility surface, electrostatics potential and conservation entropy.
The process achieves stable and high accuracies on both DNA and RNA binding proteins and illustrates how the main driving forces for nucleic acid binding are common. RNA-Puzzles is a CASP-like collective blind experiment for the evaluation of RNA three-dimensional structure prediction. The primary aims of RNA-Puzzles are to determine the capabilities and limitations of current methods of 3D RNA structure prediction based on sequence, to find whether and how progress has been made, and to illustrate whether there are specific bottlenecks that hold back the field. Dozens of puzzles have been set up with automatic assessments of the agreements with X-ray structures.
Cruz, J.A., and Westhof, E. (2011). Sequence-based identification of 3D structural modules in RNA with RMDetect. Nature methods 8, 513-521.
Miao, Z., et al. (2015) RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA, 21, 1066-1084.
Miao, Z. and Westhof, E. (2016) RBscore&NBench: a high-level web server for nucleic acid binding residues prediction with a large-scale benchmarking database. Nucleic Acids Res, 44, W562-567.
Westhof, E. (2014). Isostericity and tautomerism of base pairs in nucleic acids. FEBS letters 588, 2464-2469.


